14.6 Preparando insumos para STAN
- Lectura y adecuación de covariables
statelevel_predictors_df <-
readRDS('Recursos/Día5/Sesion1/Data/statelevel_predictors_df_dam2.rds')
## Estandarizando las variables para controlar el efecto de la escala.
statelevel_predictors_df %<>%
mutate_at(vars("luces_nocturnas",
"cubrimiento_cultivo",
"cubrimiento_urbano",
"modificacion_humana",
"accesibilidad_hospitales",
"accesibilidad_hosp_caminado"),
function(x)as.numeric(scale(x)))
head(statelevel_predictors_df,10) %>% tba()| dam2 | modificacion_humana | accesibilidad_hospitales | accesibilidad_hosp_caminado | cubrimiento_cultivo | cubrimiento_urbano | luces_nocturnas | area1 | sexo2 | edad2 | edad3 | edad4 | edad5 | anoest2 | anoest3 | anoest4 | anoest99 | tiene_sanitario | tiene_acueducto | tiene_gas | eliminar_basura | tiene_internet | piso_tierra | material_paredes | material_techo | rezago_escolar | alfabeta | hacinamiento | tasa_desocupacion | id_municipio |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 00101 | 3.6127 | -1.1835 | -1.5653 | -1.1560 | 7.2782 | 4.9650 | 1.0000 | 0.5224 | 0.2781 | 0.2117 | 0.1808 | 0.0725 | 0.2000 | 0.3680 | 0.2286 | 0.0193 | 0.0119 | 0.7946 | 0.0673 | 0.0810 | 0.6678 | 0.0033 | 0.0109 | 0.0111 | 0.3694 | 0.9247 | 0.1962 | 0.0066 | 100101 |
| 00201 | -0.0553 | 0.4449 | 0.2100 | 0.0684 | -0.0682 | -0.1511 | 0.8904 | 0.4933 | 0.2726 | 0.1849 | 0.1520 | 0.0614 | 0.3149 | 0.3022 | 0.0775 | 0.0082 | 0.1005 | 0.7220 | 0.2261 | 0.1300 | 0.9276 | 0.0664 | 0.0812 | 0.0249 | 0.1501 | 0.7975 | 0.3014 | 0.0007 | 050201 |
| 00202 | -0.3758 | 0.0000 | 0.1482 | -0.2345 | -0.2855 | -0.4234 | 0.6799 | 0.4697 | 0.2804 | 0.1895 | 0.1430 | 0.0515 | 0.3757 | 0.2405 | 0.0148 | 0.0014 | 0.1322 | 0.9230 | 0.2693 | 0.2884 | 0.9759 | 0.0625 | 0.0986 | 0.0673 | 0.0278 | 0.7140 | 0.3454 | 0.0001 | 050202 |
| 00203 | -0.9259 | 0.5732 | -0.1402 | -0.5511 | -0.3822 | -0.5612 | 0.5814 | 0.4601 | 0.2665 | 0.1733 | 0.1586 | 0.0713 | 0.3778 | 0.2463 | 0.0219 | 0.0052 | 0.2579 | 0.7602 | 0.4824 | 0.2589 | 0.9919 | 0.1937 | 0.2342 | 0.1238 | 0.0485 | 0.7104 | 0.2755 | 0.0001 | 050203 |
| 00204 | -1.3166 | 1.1111 | 0.4438 | -0.5027 | -0.3835 | -0.6042 | 0.5708 | 0.4663 | 0.2647 | 0.1683 | 0.1673 | 0.0757 | 0.3306 | 0.2402 | 0.0440 | 0.0049 | 0.1672 | 0.6375 | 0.5040 | 0.3837 | 0.9759 | 0.1403 | 0.1354 | 0.0176 | 0.0873 | 0.6737 | 0.2671 | 0.0002 | 050204 |
| 00205 | -0.7474 | 2.1155 | 1.2271 | -0.5838 | -0.3345 | -0.5909 | 0.6937 | 0.4633 | 0.2849 | 0.2107 | 0.1473 | 0.0583 | 0.2794 | 0.2821 | 0.0562 | 0.0067 | 0.3800 | 0.6596 | 0.5014 | 0.2852 | 0.9894 | 0.2309 | 0.2498 | 0.0459 | 0.1016 | 0.6751 | 0.4973 | 0.0001 | 050205 |
| 00206 | 0.5157 | -0.1468 | -0.1811 | 1.1894 | -0.1191 | -0.4022 | 0.9563 | 0.4557 | 0.2910 | 0.1814 | 0.1495 | 0.0626 | 0.3793 | 0.2815 | 0.0427 | 0.0052 | 0.1301 | 0.8817 | 0.2565 | 0.1495 | 0.9659 | 0.0629 | 0.0472 | 0.0337 | 0.0835 | 0.8027 | 0.2200 | 0.0001 | 050206 |
| 00207 | 1.7368 | -0.7648 | -0.4861 | 0.7170 | -0.0609 | 0.0042 | 0.5201 | 0.4783 | 0.2898 | 0.1675 | 0.1464 | 0.0531 | 0.3552 | 0.2901 | 0.0328 | 0.0061 | 0.2434 | 0.5775 | 0.2758 | 0.0950 | 0.9911 | 0.0717 | 0.2004 | 0.1304 | 0.0714 | 0.7778 | 0.3936 | 0.0001 | 050207 |
| 00208 | -0.5942 | 0.3212 | -0.1697 | -0.3627 | -0.3044 | -0.4750 | 0.6625 | 0.4334 | 0.2943 | 0.1875 | 0.1523 | 0.0654 | 0.3557 | 0.2486 | 0.0250 | 0.0054 | 0.1908 | 0.8251 | 0.4152 | 0.1450 | 0.9907 | 0.1458 | 0.1517 | 0.0852 | 0.0509 | 0.6897 | 0.3051 | 0.0001 | 050208 |
| 00209 | -1.5280 | 3.0192 | 1.9428 | -0.8078 | -0.4046 | -0.6423 | 0.6798 | 0.4311 | 0.2858 | 0.1687 | 0.1628 | 0.0701 | 0.3648 | 0.2645 | 0.0752 | 0.0061 | 0.1893 | 0.5760 | 0.4096 | 0.3557 | 0.9978 | 0.1097 | 0.0941 | 0.0292 | 0.1357 | 0.7680 | 0.2189 | 0.0001 | 050209 |
- Seleccionar las variables del modelo y crear matriz de covariables.
names_cov <-
c(
"dam2",
"tasa_desocupacion",
"hacinamiento",
"piso_tierra",
"luces_nocturnas",
"cubrimiento_cultivo",
"modificacion_humana"
)
X_pred <-
anti_join(statelevel_predictors_df %>% select(all_of(names_cov)),
indicador_dam1 %>% select(dam2))En el bloque de código se identifican que dominios serán los predichos.
X_pred %>% select(dam2) %>%
saveRDS(file = "Recursos/Día5/Sesion1/Data/dam_pred.rds")Creando la matriz de covariables para los dominios no observados (X_pred) y los observados (X_obs)
## Obteniendo la matrix
X_pred %<>%
data.frame() %>%
select(-dam2) %>% as.matrix()
## Identificando los dominios para realizar estimación del modelo
X_obs <- inner_join(indicador_dam1 %>% select(dam2, id_orden),
statelevel_predictors_df %>% select(all_of(names_cov))) %>%
arrange(id_orden) %>%
data.frame() %>%
select(-dam2, -id_orden) %>% as.matrix()- Calculando el n_efectivo y el \(\tilde{y}\)
D <- nrow(indicador_dam1)
P <- 3 # Ocupado, desocupado, inactivo.
Y_tilde <- matrix(NA, D, P)
n_tilde <- matrix(NA, D, P)
Y_hat <- matrix(NA, D, P)
# n efectivos ocupado
n_tilde[,1] <- (indicador_dam1$Ocupado*(1 - indicador_dam1$Ocupado))/indicador_dam1$Ocupado_var
Y_tilde[,1] <- n_tilde[,1]* indicador_dam1$Ocupado
# n efectivos desocupado
n_tilde[,2] <- (indicador_dam1$Desocupado*(1 - indicador_dam1$Desocupado))/indicador_dam1$Desocupado_var
Y_tilde[,2] <- n_tilde[,2]* indicador_dam1$Desocupado
# n efectivos Inactivo
n_tilde[,3] <- (indicador_dam1$Inactivo*(1 - indicador_dam1$Inactivo))/indicador_dam1$Inactivo_var
Y_tilde[,3] <- n_tilde[,3]* indicador_dam1$InactivoAhora, validamos la coherencia de los cálculos realizados
ni_hat = rowSums(Y_tilde)
Y_hat[,1] <- ni_hat* indicador_dam1$Ocupado
Y_hat[,2] <- ni_hat* indicador_dam1$Desocupado
Y_hat[,3] <- ni_hat* indicador_dam1$Inactivo
Y_hat <- ceiling(Y_hat)
hat_p <- Y_hat/rowSums(Y_hat)
par(mfrow = c(1,3))
plot(hat_p[,1],indicador_dam1$Ocupado)
abline(a = 0,b=1,col = "red")
plot(hat_p[,2],indicador_dam1$Desocupado)
abline(a = 0,b=1,col = "red")
plot(hat_p[,3],indicador_dam1$Inactivo)
abline(a = 0,b=1,col = "red")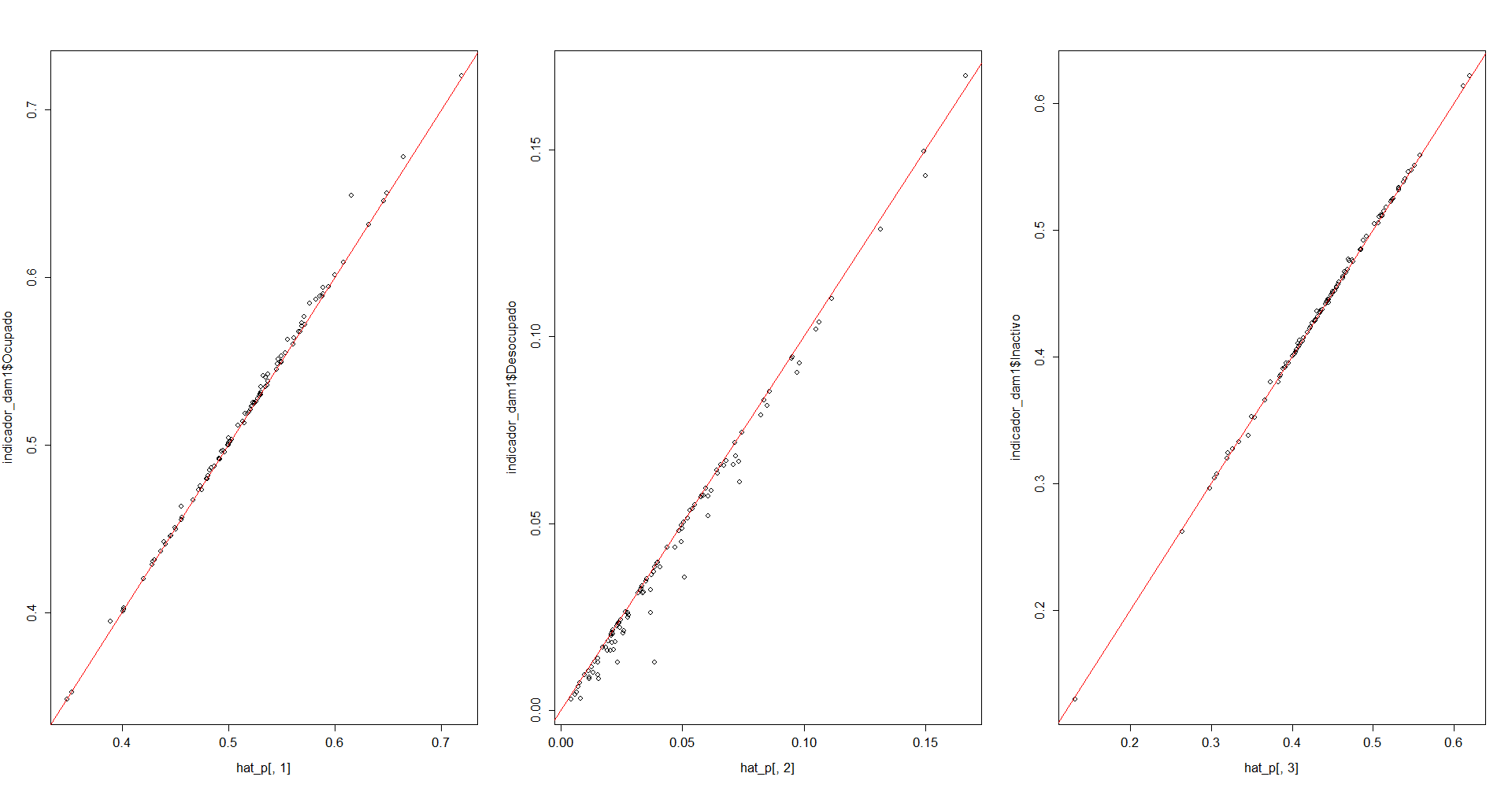
- Compilando el modelo
X1_obs <- cbind(matrix(1,nrow = D,ncol = 1),X_obs)
K = ncol(X1_obs)
D1 <- nrow(X_pred)
X1_pred <- cbind(matrix(1,nrow = D1,ncol = 1),X_pred)
sample_data <- list(D = D,
P = P,
K = K,
y_tilde = Y_hat,
X_obs = X1_obs,
X_pred = X1_pred,
D1 = D1)
library(rstan)
fit_mcmc2 <- stan(
file = "Recursos/Día5/Sesion1/Data/modelosStan/00 Multinomial_simple_no_cor.stan", # Stan program
data = sample_data, # named list of data
verbose = TRUE,
warmup = 1000, # number of warmup iterations per chain
iter = 2000, # total number of iterations per chain
cores = 4, # number of cores (could use one per chain)
)
saveRDS(fit_mcmc2,
"Recursos/Día5/Sesion1/Data/fit_multinomial_no_cor.Rds")